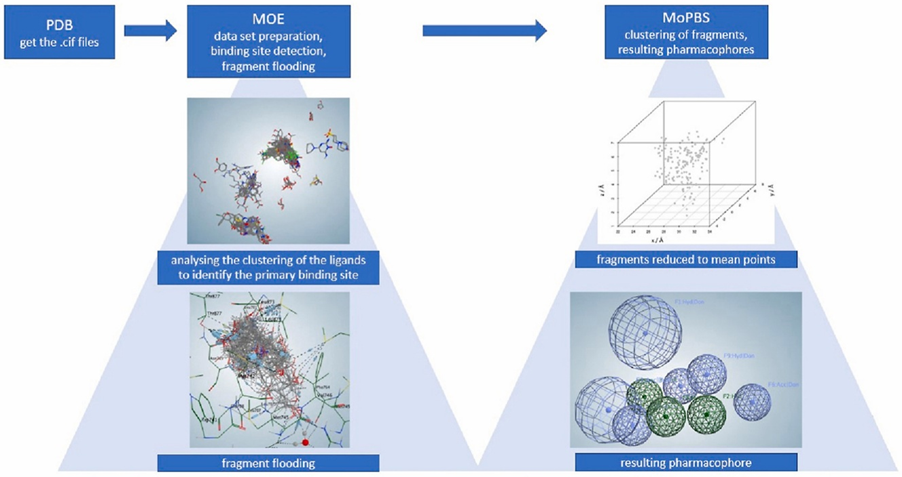
Methodology Development
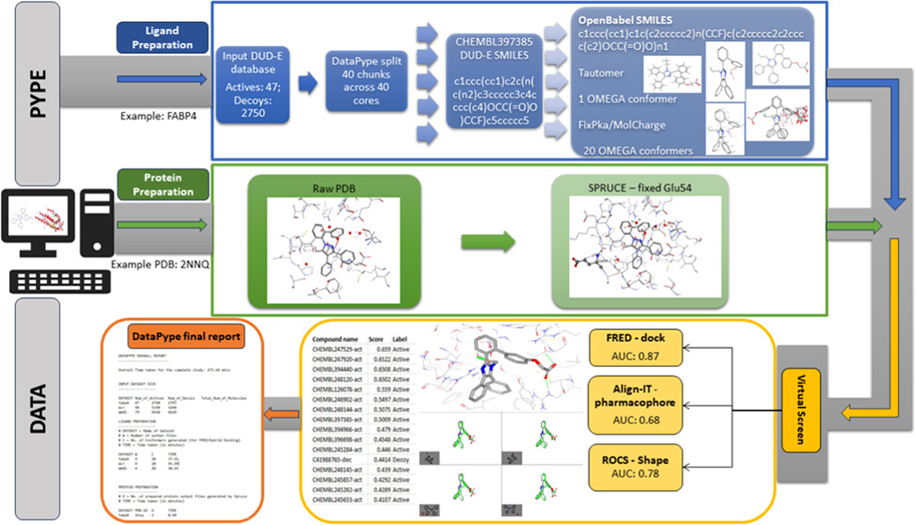
CADD application
Darren has lead a number of successful small molecule hit-finding research
programmes against various protein targets including – androgen receptor (AR),
farnesoid X receptor (FXR), peroxisome proliferating activated receptor gamma
(PPARγ), glucocorticoid receptor (GR), estrogen receptor α and β (ER), tubulin,
caspase 6, macrophage migration inhibitory factor (MIF), toll-like receptor 4/myeloid
differentiation 2 (TLR4/MD2), plasmepsins, IL-17, Viral infectivity factor (Vif),
Lipoprotein signal peptidase (LspA), cytochrome P450, SARS-CoV-2 nsp3, SARS-
CoV-2 nsp13, along with novel compounds demonstrating in vivo efficacy against
malignant pleural mesothelioma, lung cancer and sepsis.
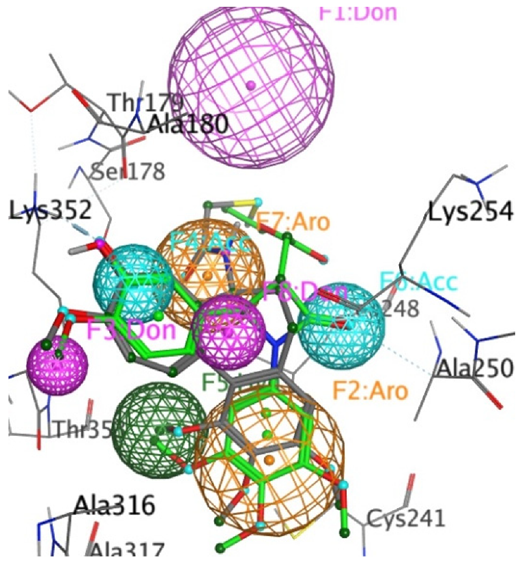
Mapping of the X-ray structure of tubulin co-crystallised with DAMA-colchicine (PDB
entry 1SA0) and the best ranked docked pose of the 3S,4R enantiomer 17g, with a
pharmacophore created by MoPBS. The ligand and protein colouring is explained in
the legend above. The pharmacophore feature colours are HBA, light blue; HBD,
pink; aromatic, orange; and hydrophobic, green [Wang, S. et al., Pharmaceuticals
2023, 16 (7). https://doi.org/10.3390/ph16071000.]